Robert B. West, Brian P. Rubin, Melinda A. Miller, Subbaya Subramanian, Gulsah Kaygusuz, Kelli Montgomery, Shirley Zhu, Robert J. Marinelli, Alessandro De Luca, Erin Downs-Kelly, John R. Goldblum, Christopher L. Corless, Patrick O. Brown, C. Blake Gilks, Torsten O. Nielsen, David Huntsman, Matt van de Rijn
| Home |
|
Home |
| Figures |
|
Figures and Tables |
| Images |
|
View the CSF1 tissue array images |
| Data |
|
View the CSF1 data |
| Methods |
|
Supplementary materials and methods |
| WebPortal |
|
Stanford Tissue Microarray Consortium Web Portal |
| Authors |
|
Authors |
| Supplemental Materials and Methods | ||
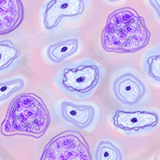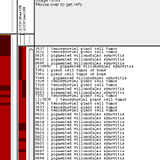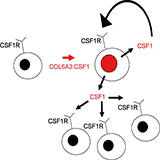
Materials and MethodsTissue microarray constructionTissue microarrays were constructed using a manual tissue arrayer (Beecher Instruments, Silver Springs, MD) following previously described techniques (17). A previously constructed set of 2 TMAs (TA38 and TA39) that represented over 50 different soft tissue tumor entities by a total of 460 600-micron cores in duplicate (a total of 920 cores) formed the initial target of ISH for the four receptor tyrosine kinases studied. These cores represent material from 421 patients with several patients having more than one specimen appearing on the arrays (8, 17). The cores were taken from soft tissue tumor samples archived at the Stanford University Medical Center Department of Pathology between 1995 and 2001. Three new TMAs (TA117, TA137 and TA153) were constructed for the purpose of this study and the cases represented on them are shown in Supplemental Table 1. TA137 has 9 cases of TGCT from TA38. TA39, and TA117 were used for the initial receptor tyrosine kinase screening, representing 507 cases of soft tissue tumors. TA137 and TA153 were used for further FISH and ISH analysis of TGCT and PVNS. RNA in situ hybridizationIn situ hybridization of TMA sections was performed based on a protocol published previously (8). Briefly, digoxigenin (DIG)-labeled sense and anti-sense RNA probes are generated by PCR amplification of 400 to 600 bp products with the T7 promoter incorporated into the primers. The primer sequences are given in Supplemental Table 2 on this web site. In vitro transcription was performed with a DIG RNA-labeling kit and T7 polymerase according to the manufacturer's protocol (Roche Diagnostics, Indianapolis, IN). 5 micron thick paraffin sections were deparaffinized in xylene and were hydrated in graded concentrations of ethanol for 5 minutes each. Sections were then incubated with 1% hydrogen peroxide, followed by digestion in 10ug/ml of proteinase K at 37°C for 30 minutes. Sections were hybridized overnight at 55oC with either sense or antisense riboprobes at 200ng/ml dilution in mRNA hybridization buffer (Dako). The following day, sections were washed in 2xSSC and incubated with 1:35 dilution of RNase A cocktail (Ambion, Austin, TX) in 2xSSC for 30minutes at 37°C. Next, sections were washed twice in 2X SSC/50% formamide, followed by a wash in 0.08X SSC at 50 °C. Biotin blocking reagents (Dako) were applied to the section to block the endogenous biotin. For signal amplification, a HRP-conjugated rabbit anti-DIG antibody (Dako) was used to catalyze the deposition of biotinyl tyramide, followed by secondary streptavidin complex conjugated with horseradish peroxidase (GenPoint kit; Dako). The final signal was developed with Vector VIP substrate kit, and the tissues were counterstained in hematoxylin for 15 seconds. TA38, 39, 137, 153, and 117 were stained with oligo dT to assess preservation of tissue RNA using the INFORM in situ hybridization Oligo dT control probe (Ventana Medical Systems, Tuscon, AZ, USA) In situ hybridization scoring was based on recognizing a strong dot-like staining pattern associated with cells. Only cores that had RNA as detected by an oligo dT probe were scored. A score of negative ("0") was given for less than 1 dot-like stain per 2-3 lesional cells. A score of weak positive ("1") was given for 1 dot-like stain per 1 to 3 lesional cells. A score of strong positive ("2") was given for greater than 1 dot-like stain per lesional cell. ImmunohistochemistryA primary antibody towards CD163 (Novocastra Laboratories, New Castle Upon Tyne, UK) was used. Briefly, 4 micron serial sections were cut from the TMA blocks, deparaffinized in xylene, and hydrated in a graded series of alcohol. The slides were pretreated with citrate buffer and a microwave step. Staining was then performed using the DAKO EnVision+ System and the Vector VIP substrate kit. Digital images of all cores stained by H&E or immunohistochemistry were collected using the BLISS system from Bacuslabs (Lombard IL, http://bacuslabs.com). For double staining, in situ hybridization was performed, followed by the immunostaining. Combined ISH and ImmunohistochemistryFor double staining, in situ hybridization was performed first, followed by the immunostaining. A streptavidin Alexa Fluor 594 conjugate (S-32356, Invitrogen Corp., Carlsbad, CA, USA) diluted at 1:500 was used to visualize the ISH probe for CSF1. The slide was imaged using a Nikon E1000 microscope with UV-2E DAPI and Texas Red HYQ filter cubes (Nikon Instruments Inc., Melville, NY, USA) fitted with a CoolSNAP K4 (Photometrics, Tucson, AZ, USA) camera. Subsequently, the cover-slip was removed and the slide was re-stained with anti-CD163 antibody (Novocastra, diluted 1:100) using the DAKO EnVision+ System and the Vector VIP substrate kit and counterstained with hematoxylin. The slide was imaged again using bright-field and RGB color filters. The fluorescence image was aligned to and superimposed on the bright-field image by selecting the DAPI channel using Photoshop CS Version 8 (Adobe Systems, Inc., San Jose, CA, USA). For the final image the DAPI channel was removed, leaving the Texas Red channel superimposed on the bright field image. Fluorescence In Situ HybridizationSix micron thick sections of the TMA slides were pre-treated as described elsewhere (18). Metaphases and metaphase slides were produced using standard methods. Locus specific FISH analysis was performed using the following bacterial artificial chromosomes (BACs) from the Human BAC Library RPCI-11 (BACPAC Resources Centre, Children's Hospital Oakland Research Institute) unless otherwise noted. Listed centromeric to telomeric: 354C7, 19F3 and 96F24 (chromosome 1) and 155J6, 205L13, 585E12, 97L10, CTD-2344F21 (CITB Human D BAC library, Invitrogen), 279M4, 258O3, 497D24 (chromosome 2). BACs were directly labeled with either Spectrum Green or Spectrum Orange (Vysis, Downer’s Grove, IL). The chromosomal locations of all BACs were validated using normal metaphases (results not shown). Probe labelling and FISH was performed using Vysis reagents according to manufacturer's protocols (Vysis, IL). Slides were counterstained with 4,6-diamidino 2-phenylindole (DAPI) for microscopy. For all slides, FISH signals and patterns were identified on a Zeiss Axioplan epifluorescent microscope. Signals were interpreted manually and images were captured using Metasystems Isis FISH imaging software (MetaSystems Group, Inc. Belmont, MA). A cutoff of equal to or greater than 2 breaks per 100 nuclei was selected for a positive score based on examining 230 other soft tissue tumors. Efficiency of the break apart FISH probes on TMAs were previously demonstrated with the t(X;18) in synovial sarcomas (19). Combined Fluorescence In Situ Hybridization and immunohistochemistryCombined FISH and immunohistochemistry was performed using the pretreatment steps of the standard immunohistochemistry, followed by application of the antibody, followed by the hybridization with the FISH probes. BACs were directly labeled with either Spectrum Green or Spectrum Orange (Vysis, Downer's Grove, IL). The immunohistochemistry was performed with a VECTASTAIN Elite ABC Kit (PK-6105, Vector Labs., Burlingame, CA, USA) and labeled with a streptavidin Alexa Fluor 647 conjugate (S-32357, Invitrogen Corp., Carlsbad, CA, USA). Gene arrayTumors were collected from three academic institutions (Vancouver General Hospital, University of Washington Medical Center, and Stanford University Medical Center) with institutional IRB approval. After resection, a representative sample was quickly frozen and stored at -80°C. Prior to processing, frozen sections of the tissue were cut and histologically examined to ensure that the tissue represented the diagnostic entity. Only cases with classic histologic findings were used. 8 cases of PVNS and 7 cases of TGCT were compared to 6 cases of solitary fibrous tumor (SFT) and 7 cases of fibromatosis (DTF). In the cases of SFT and DTF, all cases except two (STT3237 and STT3524) have been previously published (20). 42,000 spot cDNA microarrays were used to measure the relative mRNA expression levels in the tumors. The details of isolating mRNA, labeling and hybridizing are described elsewhere (21). The raw data files are available at Stanford Microarray Database (http://genome-www5.stanford.edu), the filtered data used for the manuscript are available at the accompanying web site http://tma.stanford.edu/tma_portal/csf1/ in a searchable format. Data was filtered using the following criteria: only cDNA spots with a ratio of signal over background of at least 1.5 in both the Cy3 and the Cy5 channel were included; only cDNA spots that fullfill these criteria on at least 70% arrays were included; and only cDNAs were selected that had an absolute value at least four times greater in at least two arrays than the geometric mean. Data was evaluated with unsupervised hierarchical clustering and significance analysis of microarrays (SAM) (22). References(References are numbered to match the paper.) |
||
|
|
||
|
|
||