Selecting Score Combination Rules – Part 1
This section represents what is perhaps the most important part
of the TMA-Combiner. At first glance, one may think that combining
scores by simple averaging would be sufficient. However, when it
comes to combining stained and scored IHC replicate cores, the situation
is not that simple! One needs to take a step back and carefully
consider the significance of what the combination process is doing
to these IHC-stained replicate cores. Issues such as core sampling
biases, antibody staining properties, and number of replicate cores
weigh considerably in this process.
We discuss the importance of these considerations and why we
constructed the score combination rules shown below, in our publication,
so we won't repeat it here. We will, however, go into greater detail
on the mechanics of the score combination process, particularly on how
each rule determines the outcome score from the input score pool of
replicate cores.
First off, it is important to establish the scoring system that
is introduced earlier in the TMA-Deconvoluter walkthrough. The TMA-Combiner
can handle quantitative scoring systems, which we describe in greater
detail here. However, we
use a discrete integer scoring system, so the TMA-Combiner defaults to
that, as shown in the score key below:
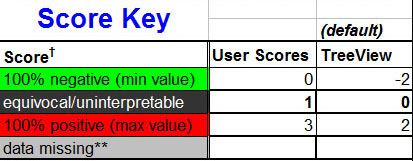
† Background colors indicate how they
would appear in TreeView
Note that scores are either user scores or TreeView scores. This
is dependent on the type of output selected for the TMA-Deconvoluter
- PCL files use TreeView-compatible scores, whereas K-M files use unconverted,
user scores. User scores might also be present if the user
generated the input files from other sources or with methods other
than the TMA-Deconvoluter.
In order for the TMA-Combiner to apply the score combination rules
correctly, the user will need to indicate whether or not user
scores are present in the input files. Since the PCL file format is the native format for
the TMA-Combiner, the TMA-Combiner will assume by default that the input files contain
TreeView-compatible scores. Thus, if the user is combining scores based on the user's
scoring system, the user should select the option below:

**In the case of missing datapoints, while the default is a blank cell, the user
may use a number or symbol (such as "X") instead. This should be indicated in
the Score Key, like in the example below:
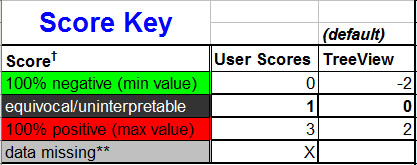
One other, very important consideration with scores – if the
user is planning to combine multiple TMAs together into a single
file, the user should ensure that every file in the entire dataset
consists entirely of user scores or of TreeView-compatible scores (i.e. the
constituent files comprising the whole dataset to be combined should contain only
one type of score, not both).
Otherwise, score combination might not be performed correctly and may produce
errors. The easiest way to fix this is to use the
TMA-Deconvoluter's Score Conversion Utility to convert the scores. For more
information on this, consult the TMA-Deconvoluter
documentation.
back to top
Back to Step 1
Step 3 - Score Combination Rules, Part 2
Return to the walkthrough
overview page.
|

