 | Discovery of Molecular Subtypes in Leiomyosarcoma through Integrative Molecular Profiling
Andrew H Beck, Cheng-Han Lee, Daniela M Witten, Briana C Gleason, Badreddin Edris, Inigo Espinosa, Shirley Zhu, Rui Li, Kelli D Montgomery, Robert J Marinelli, Robert Tibshirani, Trevor Hastie, David M Jablons, Brian P Rubin, Christopher D Fletcher, Robert B West, Matt van de Rijn |
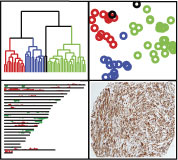
| Welcome to the web supplement to the paper:Discovery of Molecular Subtypes in Leiomyosarcoma through Integrative Molecular Profiling Oncogene advance online publication 9 November 2009; doi: 10.1038/onc.2009.381 | Abstract | | Leiomyosarcoma (LMS) is a soft tissue tumor with a significant degree of morphologic and molecular heterogeneity. We used integrative molecular profiling to discover and characterize molecular subtypes of LMS. Gene expression profiling was performed on 51 LMS samples. Unsupervised clustering showed three reproducible LMS clusters. Array comparative genomic hybridization (aCGH) was performed on 20 LMS samples and showed that the molecular subtypes defined by gene expression showed distinct genomic changes. Tumors from the 'muscle-enriched' cluster showed significantly increased copy number changes (P=0.04). A majority of the muscle-enriched cases showed loss at 16q24, which contains Fanconi anemia, complementation group A, known to have an important role in DNA repair, and loss at 1p36, which contains PRDM16, of which loss promotes muscle differentiation. Immunohistochemistry (IHC) was performed on LMS tissue microarrays (n=377) for five markers with high levels of messenger RNA in the muscle-enriched cluster (ACTG2, CASQ2, SLMAP, CFL2 and MYLK) and showed significantly correlated expression of the five proteins (all pairwise P<0.005). Expression of the five markers was associated with improved disease-specific survival in a multivariate Cox regression analysis (P<0.04). In this analysis that combined gene expression profiling, aCGH and IHC, we characterized distinct molecular LMS subtypes, provided insight into their pathogenesis, and identified prognostic biomarkers. |
|
